-Search query
-Search result
Showing all 44 items for (author: tuck & ot)

EMDB-43613: 
Structure of HamAB apo complex from the Escherichia coli Hachiman defense system
Method: single particle / : Tuck OT, Doudna JA

EMDB-43615: 
Structure of HamB-DNA complex, conformation 1, from the Escherichia coli Hachiman defense system
Method: single particle / : Tuck OT, Doudna JA

EMDB-43616: 
Structure of HamB-DNA complex, conformation 2, from the Escherichia coli Hachiman defense system
Method: single particle / : Tuck OT, Doudna JA

EMDB-43643: 
Structure of HamA(E138A,K140A)B-plasmid DNA complex from the Escherichia coli Hachiman defense system
Method: single particle / : Tuck OT, Hu JJ, Doudna JA

EMDB-29561: 
Cryo-EM structure of Cas1:Cas2-DEDDh:PAM-deficient prespacer complex
Method: single particle / : Skopintsev P, Tuck OT, Soczek KM, Doudna J

EMDB-29562: 
Cryo-EM structure of Cas1:Cas2-DEDDh:PAM-containing prespacer complex
Method: single particle / : Skopintsev P, Tuck OT, Soczek KM, Doudna J

EMDB-29563: 
Cryo-EM structure of Cas1:Cas2-DEDDh:half-site integration complex
Method: single particle / : Skopintsev P, Tuck OT, Soczek KM, Doudna J

EMDB-29564: 
Cryo-EM structure of Cas1:Cas2-DEDDh:half-site integration complex linear CRISPR repeat conformation
Method: single particle / : Skopintsev P, Tuck OT, Soczek KM, Doudna J

EMDB-29565: 
Cryo-EM structure of Cas1:Cas2-DEDDh:half-site integration complex bent CRISPR repeat conformation
Method: single particle / : Skopintsev P, Tuck OT, Soczek KM, Doudna J

EMDB-29396: 
Antibody vFP53.02 in complex with HIV-1 envelope trimer BG505 DS-SOSIP
Method: single particle / : Wang S, Kwong PD
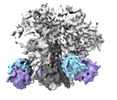
EMDB-29836: 
vFP52.02 Fab in complex with BG505 DS-SOSIP Env trimer
Method: single particle / : Gorman J, Kwong PD

EMDB-29880: 
Cryo-EM structure of vFP49.02 Fab in complex with HIV-1 Env BG505 DS-SOSIP.664 (conformation 1)
Method: single particle / : Changela A, Gorman J, Kwong PD

EMDB-29881: 
Cryo-EM structure of vFP49.02 Fab in complex with HIV-1 Env BG505 DS-SOSIP.664 (conformation 2)
Method: single particle / : Changela A, Gorman J, Kwong PD

EMDB-29882: 
Cryo-EM structure of vFP49.02 Fab in complex with HIV-1 Env BG505 DS-SOSIP.664 (conformation 3)
Method: single particle / : Changela A, Gorman J, Kwong PD

EMDB-29905: 
vFP48.02 Fab in complex with BG505 DS-SOSIP Env trimer
Method: single particle / : Gorman J, Kwong PD

PDB-8fr6: 
Antibody vFP53.02 in complex with HIV-1 envelope trimer BG505 DS-SOSIP
Method: single particle / : Wang S, Kwong PD

PDB-8g85: 
vFP52.02 Fab in complex with BG505 DS-SOSIP Env trimer
Method: single particle / : Gorman J, Kwong PD

PDB-8g9w: 
Cryo-EM structure of vFP49.02 Fab in complex with HIV-1 Env BG505 DS-SOSIP.664 (conformation 1)
Method: single particle / : Changela A, Gorman J, Kwong PD

PDB-8g9x: 
Cryo-EM structure of vFP49.02 Fab in complex with HIV-1 Env BG505 DS-SOSIP.664 (conformation 2)
Method: single particle / : Changela A, Gorman J, Kwong PD

PDB-8g9y: 
Cryo-EM structure of vFP49.02 Fab in complex with HIV-1 Env BG505 DS-SOSIP.664 (conformation 3)
Method: single particle / : Changela A, Gorman J, Kwong PD

PDB-8gas: 
vFP48.02 Fab in complex with BG505 DS-SOSIP Env trimer
Method: single particle / : Gorman J, Kwong PD

EMDB-24299: 
SARS-CoV-2 spike in complex with the S2D106 neutralizing antibody Fab fragment (local refinement of the RBD and S2D106)
Method: single particle / : Park YJ, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

EMDB-24300: 
SARS-CoV-2 spike in complex with the S2D106 neutralizing antibody Fab fragment
Method: single particle / : Park YJ, Veesler D

EMDB-24301: 
SARS-CoV-2 spike glycoprotein ectodomain in complex with the S2H97 neutralizing antibody Fab fragment
Method: single particle / : Park YJ, Veesler D

PDB-7r7n: 
SARS-CoV-2 spike in complex with the S2D106 neutralizing antibody Fab fragment (local refinement of the RBD and S2D106)
Method: single particle / : Park YJ, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

EMDB-30915: 
Apo spike protein of SARS-CoV2
Method: single particle / : Liu Y, Soh WT, Kishikawa J, Hirose M, Kato T, Okada M, Arase H

EMDB-30916: 
SARS-CoV2 spike protein with Fab fragment of enhancing antibody 8D2
Method: single particle / : Liu Y, Soh WT, Kishikawa J, Hirose M, Kato T, Okada M, Arase H

EMDB-30917: 
SARS-CoV2 spike protein with Fab fragment of enhancing antibody (8D2)-1
Method: single particle / : Liu Y, Soh WT, Kishikawa J, Hirose M, Kato T, Okada M, Arase H

EMDB-30918: 
SARS-CoV2 spike protein with Fab fragment of enhancing antibody (8D2)-2
Method: single particle / : Liu Y, Soh WT, Kishikawa J, Hirose M, Kato T, Okada M, Arase H

EMDB-30919: 
SARS-CoV2 spike protein with Fab fragment of enhancing antibody (8D2)-3
Method: single particle / : Liu Y, Soh WT, Kishikawa J, Hirose M, Kato T, Okada M, Arase H

EMDB-30920: 
SARS-CoV2 spike protein with Fab fragment of enhancing antibody (8D2)-4
Method: single particle / : Liu Y, Soh WT, Kishikawa J, Hirose M, Kato T, Okada M, Arase H

EMDB-30921: 
SARS-CoV2 spike protein with Fab fragment of enhancing antibody (2940)-1
Method: single particle / : Liu Y, Soh WT, Kishikawa J, Hirose M, Kato T, Okada M, Arase H

EMDB-30922: 
SARS-CoV2 spike protein with Fab fragment of enhancing antibody (2940)-2
Method: single particle / : Liu Y, Soh WT, Kishikawa J, Hirose M, Kato T, Okada M, Arase H

PDB-7dzw: 
Apo spike protein from SARS-CoV2
Method: single particle / : Liu Y, Soh WT, Li S, Kishikawa J, Hirose M, Kato T, Standley D, Okada M, Arase H

PDB-7dzx: 
Spike protein from SARS-CoV2 with Fab fragment of enhancing antibody 8D2
Method: single particle / : Liu Y, Soh WT, Li S, Kishikawa J, Hirose M, Kato T, Standley D, Okada M, Arase H

PDB-7dzy: 
Spike protein from SARS-CoV2 with Fab fragment of enhancing antibody 2490
Method: single particle / : Liu Y, Soh WT, Li S, Kishikawa J, Hirose M, Kato T, Standley D, Okada M, Arase H

EMDB-21140: 
Cryo-EM structure of a dimer of undecameric human CALHM2
Method: single particle / : Syrjanen JL, Chou TH, Furukawa H

EMDB-21141: 
Cryo-EM structure of human CALHM2
Method: single particle / : Syrjanen JL, Chou TH, Furukawa H

EMDB-21142: 
Cryo-EM structure of an undecameric chicken CALHM1 and human CALHM2 chimera
Method: single particle / : Syrjanen JL, Chou TH, Furukawa H

EMDB-21143: 
Cryo-EM structure of octameric chicken CALHM1
Method: single particle / : Syrjanen JL, Chou TH, Furukawa H

PDB-6vai: 
Cryo-EM structure of a dimer of undecameric human CALHM2
Method: single particle / : Syrjanen JL, Chou TH, Furukawa H

PDB-6vak: 
Cryo-EM structure of human CALHM2
Method: single particle / : Syrjanen JL, Chou TH, Furukawa H

PDB-6val: 
Cryo-EM structure of an undecameric chicken CALHM1 and human CALHM2 chimera
Method: single particle / : Syrjanen JL, Chou TH, Furukawa H

PDB-6vam: 
Cryo-EM structure of octameric chicken CALHM1
Method: single particle / : Syrjanen JL, Chou TH, Furukawa H
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
